Parallel Read and Write
NumPy Slicing Syntax
PnetCDF-python datasets re-use the
numpyslicing syntax to read and write to the file. Slice specifications are translated directly to PnetCDF-C style of subarray selection, i.e. using index arrays of “start, count, stride”. The following slicing arguments are recognized:
Indices (var[1,5])
Slices (i.e. [:] or [0:10])
An empty tuple (()) to retrieve all data
Multiple indexing (e.g. var[1][5]) is NOT SUPPORTED in write
The operational mode (collective/independent) is dependent on the current file mode status.
f.enddef() # Exit define mode var = f.variables['var'] buff = np.zeros(shape = (10, 50), dtype = "i4") # put values to the entire variable var[:] = buff # read the top-left 10*10 corner from variable var print(var[:10, :10])
Method Call of put_var()/get_var()
Using specific method calls to perform I/O is particularly useful in multi-processing programs.
Variable.put_var()requires data as a mandatory argument, which serves as a write buffer that stores values to be written.Variable.get_var()requires buff as a mandatory argument, which serves as a read buffer that stores values to be read. The behavior ofVariable.put_var()andVariable.get_var()varies depending on the pattern of provided optional arguments - start, count, stride, and imap. The suffix _all indicates this is collective I/O in contrast to independent I/O (without _all).
- Read from netCDF variables
For reading, the behavior of
Variable.get_var()depends on the following provided input parameter pattern:buff - Read an entire variable
buff, start - Read a single data value
buff, start, count - Read an array of values
buff, start, count, stride - Read a subarray of values
buff, start, count, imap, buff - Read a mapped array of values
where start, count and stride represent a corner, a vector of edge lengths, and a stride vector respectively. Together, they specify a subarray section to read from in a netCDF variable as illustrated in the diagram below. By default, the method returns a multidimensional numpy array in the shape of (count[0], … count[n-1]).
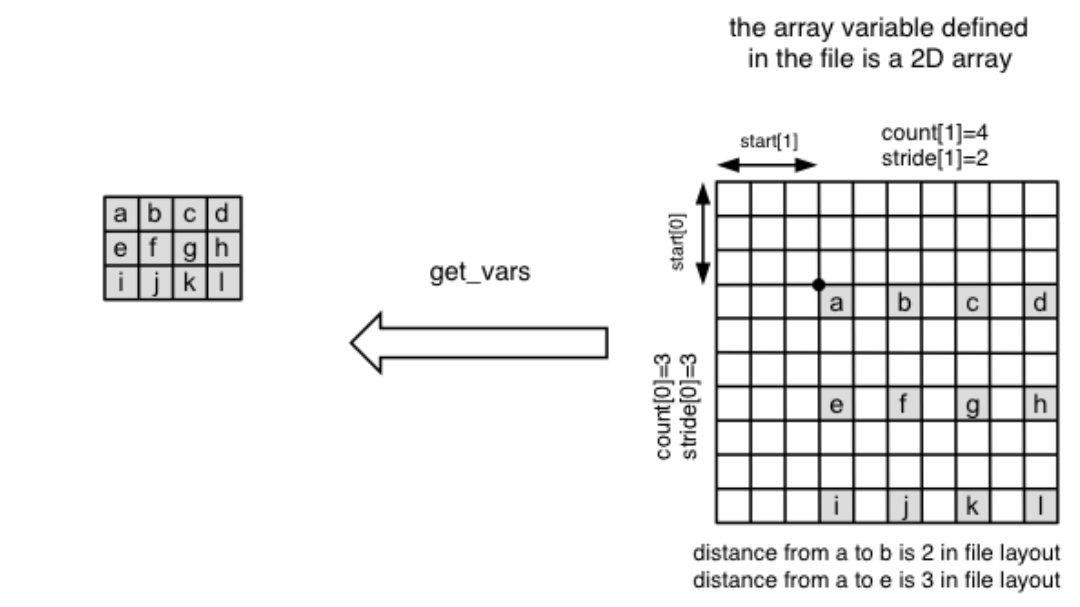
Here’s a python example:
# Collectively read from a subarray of a variable buf = var.get_var_all(start = [0, 0], count = [5, 25], stride = [2,2]) # Independently read from a subarray of a variable f.end_indep() buf = var.get_var(start = [0, 0], count = [5, 25], stride = [2,2])
For full example program, see
examples/get_var.py.- Write to netCDF variables
For writing, the behavior of
Variable.put_var()depends on the following provided input parameter pattern:data - Write an entire variable
data, start - Write a single data value (a single element)
data, start, count - Write an array of values
data, start, count, stride - Write a subarray of values
data, start, count, imap - Write a mapped array of values
where start, count and stride represent a corner, a vector of edge lengths, and a stride vector respectively. Together, they specify a subarray section to write to for a netCDF variable as illustrated in the diagram below. Note that the buffer array (the numpy array to write) can take any shape as long as the total size is matched with count.
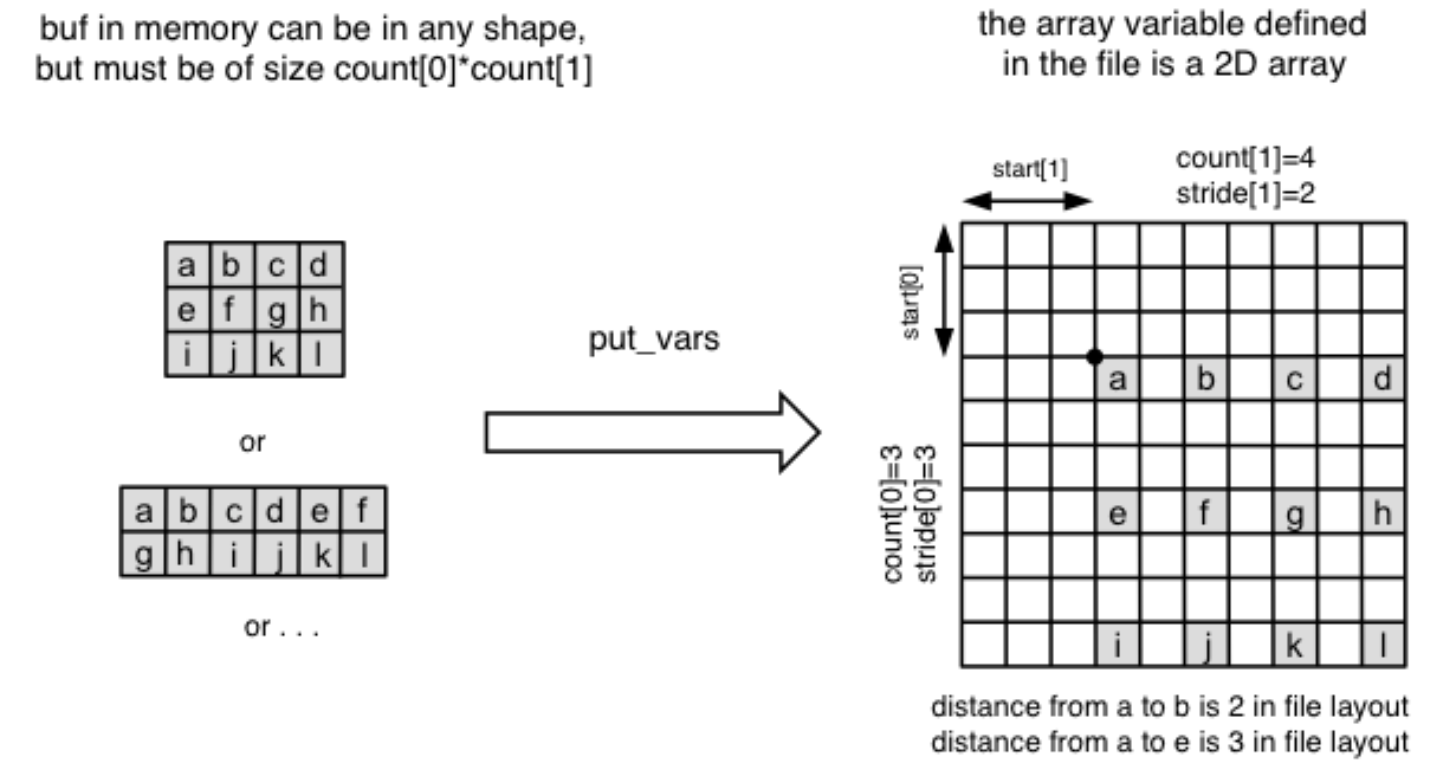
Here’s a python example:
# Collectively write to a subarray of a variable buff = np.zeros(shape = (5, 25), dtype = "i4") var.put_var_all(buff, start = [0, 0], count = [5, 25], stride = [2,2]) # Independently write to a subarray of a variable f.end_indep() var.put_var(buff, start = [0, 0], count = [5, 25], stride = [2,2])
For the full example program, see
examples/put_var.pyandexamples/collective_write.py.